Chemogenetic kinetics (2): Extinction
Biologically uninformed theorists failed to link light-activated energy-dependent “conditions of life” to biophysically constrained viral latency. That led them to ignore the consequences of virus-driven degradation of messenger RNA in species from microbes to humans.
Their ignorance led to recent claims about the discovery of a model, which places Charles Darwin’s prescient claims about natural selection for food energy-dependent codon optimality into the context of the physiology of reproduction and all biodiversity on Earth. Simply put, finches and other organisms must eat or they do not reproduce and their species becomes extinct.
Only the words used to describe what is known about natural selection for the food energy-dependent physiology of reproduction, reproductive fitness, and healthy longevity have changed since Darwin’s time.
See for example: A cell fitness selection model for neuronal survival during development 9/12/19
…in this model, a heterogeneous code of intrinsic cell fitness in neighboring neurons provides differential competitive advantage resulting in the selection of cells with higher capacity to survive and functionally integrate into neural networks.
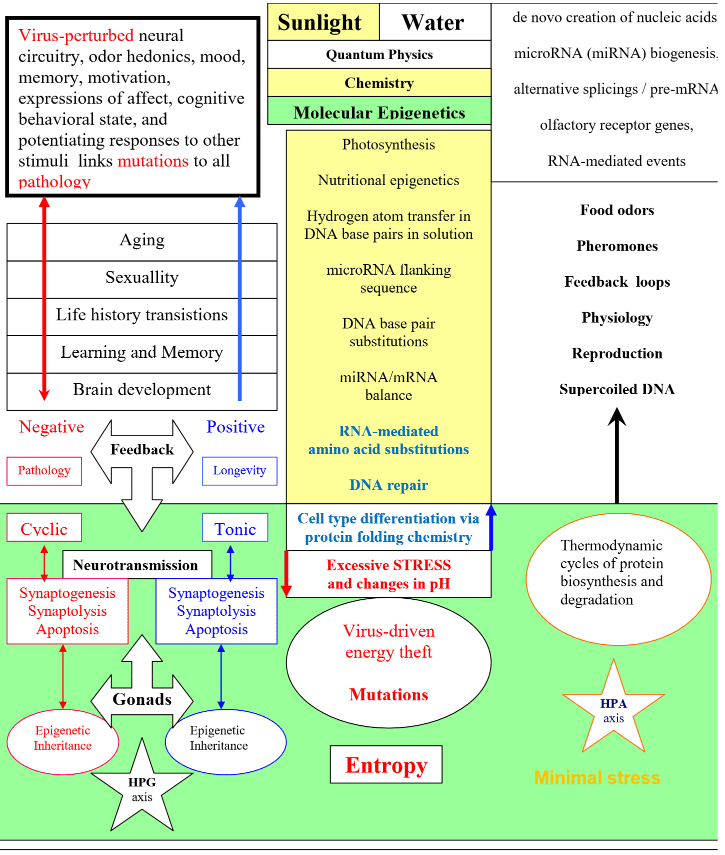
Energy-dependent autophagy links their so-called “selection model” from Darwin’s theory of natural selection via the heterogenous code of intrinsic cell fitness, which is biophysically constrained by cell type differentiation via the Creation of sunlight, water, and supercoiled DNA.
That fact was reported as: Nervous system discovery overturns previous theory 9/12/19
Here is what was included in the previous theory: “chance determines” and a “random process”
Excerpt 1)
The results indicate that the longstanding neurotrophic theory, which states that chance determines which cells will form the nervous system, needs to be revised.
Excerpt 2)
Researchers have previously believed this to be a random process in which all cells have an equal chance of survival. However, researchers from Karolinska Institute have now publishing a study in Nature Communications showing that cell death instead appears to be controlled by a mechanism that weeds out the less fit cells.
Autophagy …is the natural, regulated mechanism of the cell that removes unnecessary or dysfunctional components.[3] It allows the orderly degradation and recycling of cellular components.[4][5]
Serious scientists never believed that autophagy was a chance-determined random process. Pseudoscientists placed the previous theory into the ridiculous context of mutation-driven evolution, which made no sense after publication of Olfaction Warps Visual Time Perception (2018).
These findings point to a neuronal network in the right temporal cortex that executes flexible temporal filtering of upstream visual inputs based on olfactory information. Moreover, they collectively indicate that the very process of sensory integration at the stage of object processing twists time perception, hence casting new insights into the neural timing of multisensory events.
Wen Zhou’s group linked the Creation of sunlight and water to our visual perception of mass and energy via multisensory events and the physiology of reproduction.
Rebecca K. Skalsky’s group linked Reduced Expression of Brain-Enriched microRNAs in Glioblastomas Permits Targeted Regulation of a Cell Death Gene 9/2/11 to stories still told by biologically uninformed theorists who have failed to link the light-activated Creation and assembly of the microRNA-RNA-peptide nanocomplex to the creation of supercoiled DNA and prevention of viral replication.
See: Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs 5/9/14
…if viruses indeed evolved to avoid miRNA binding by the selection of point mutants that destroy potential miRNA binding sites, then they would be more likely to be resistant to the pattern of miRNA expression seen in their normal target tissues and less likely to be resistant to the novel miRNAs seen in a cell type, such as 293T cells, that they normally would never encounter in vivo. Nevertheless, we observed no evidence that cellular miRNAs are able to significantly repress the replication of wild-type viruses.
This story is that they ‘…observed no evidence…” Darwin observed the evidence of Creation, but their claim was made a few years before the technology became available that links the measurement of fluorescence in cell types to microRNA-mediated biophysically constrained viral latency and the cure for all pathology. The cures was patented as RNA-Guided Human Genome Engineering
5. Repetitive elements or endogenous viral elements can be targeted with engineered Cas+gRNA systems in microbes, plants, animals, or human cells to reduce deleterious transposition or to aid in sequencing or other analytic genomic/transcriptomic/proteomic/diagnostic tools (in which nearly identical copies can be problematic).
See also: Special Issue “Non-Coding RNAs in Viral Infections” 9/21/18
Non-coding RNAs (ncRNAs) are key regulators in anti-viral responses and play diverse roles in various aspects of infection, including virus replication, persistence, and pathogenesis. In addition to host ncRNAs that are influenced and/or utilized during infection, both DNA and RNA viruses produce regulatory ncRNAs. These molecules can act to modulate gene expression, such as microRNAs encoded by several DNA viruses, or control RNA stability or epigenetic processes, such as some viral long ncRNAs. Ongoing questions include how viral ncRNAs contribute to immune evasion, whether ncRNAs act as virulence factors, how viruses hijack host ncRNAs, and how both viral and host ncRNAs coordinate to benefit virus replication.
MicroRNAs are non-coding RNAs (ncRNAs). Light-activated microRNA biogenesis prevents virus replication and prevention is required to link microRNA biogenesis to the genesis of all biodiversity via naturally occurring RNA interference which is the key to successful autophagy.
See also: Functional Interplay between RNA Viruses and Non-Coding RNA in Mammals 1/14/19
…we discuss direct and indirect RNA interactions occurring between RNA viruses or retroviruses and host non-coding transcripts upon infection. In addition, we review RNA virus derived non-coding RNAs affecting immunological and metabolic pathways of the host cell typically to provide an advantage to the virus. The relatively few known examples of virus–host RNA interactions suggest that many more await discovery.
Claims about pending discoveries are often used to obfuscate claims that already fully support the paradigm shift that occurred many years ago. Then, suddenly we see the newsworthy report, Nervous system discovery overturns previous theory 9/12/19
But wait, McEwen et al., (1964) Dependence of RNA synthesis in isolated thymus nuclei on glycolysis, oxidative carbohydrate catabolism and a type of “oxidative phosphorylation” wrote:
The synthesis of RNA in isolated thymus nuclei is ATP dependent.
The synthesis of ATP is energy-dependent and nothing good in life happens outside the context of energy-dependent links from the Creation of sunlight and water to the Creation of ATP, microRNA biogenesis and the Creation of RNA, which biophysically constrains viral latency.
For an example of something bad that happens in the context of the virus-driven degradation of messenger RNA, see:
Herpes Simplex Virus 1 Deregulation of Host MicroRNAs 11/23/18
Viruses utilize microRNAs (miRNAs) in a vast variety of possible interactions and mechanisms, apparently far beyond the classical understanding of gene repression in humans. Likewise, herpes simplex virus 1 (HSV-1) expresses numerous miRNAs and deregulates the expression of host miRNAs. Several HSV-1 miRNAs are abundantly expressed in latency, some of which are encoded antisense to transcripts of important productive infection genes, indicating their roles in repressing the productive cycle and/or in maintenance/reactivation from latency. In addition, HSV-1 also exploits host miRNAs to advance its replication or repress its genes to facilitate latency. Here, we discuss what is known about the functional interplay between HSV-1 and the host miRNA machinery, potential targets, and the molecular mechanisms leading to an efficient virus replication and spread.
Discussion of what is known has again been linked to the refutation of all ridiculous theories. The continuing saga of how ignorance has prevented many people from linking viruses to all pathology can be viewed in the context of evidence:
Herpes Viruses and Senile Dementia: First Population Evidence for a Causal Link 6/19/18
The ignorance can also be viewed in the context of disease prevention for comparison to The maddening saga of how an Alzheimer’s ‘cabal’ thwarted progress toward a cure for decades 6/25/19
The virus-driven degradation of messenger RNA was linked to Alzheimer’s disease in the context of how a genetically-engineered yeast was delivered to a targeted human population in Quantico (2006).
The author’s thoughts about the next viral pandemic were placed into the context of comic relief in: Sci-fi author Greg Bear tells Jon about the not-so-distant future of technology and helping Homeland Security (2007 video)
See also:
Here we identify hundreds of RNA G-quadruplex (rG4) candidates in microRNAs (miRNAs), characterize the miRNA structure and miRNA-mRNA interactions on several mammalian-conserved miRNAs, and reveal the formation of rG4s in miRNAs. Notably, we study the effect of these rG4s in cells and uncover the role of rG4s in miRNA-mediated post-transcriptional regulation.
They used improved ILP and biophysical assays to link energy-dependent changes in SNPs from base pairs to our diet-driven formation of rG4 structures and the effect of miRNAs on RNA-mediated miRNA-mRNA interactions, which typically prevent all the diseases linked to extinctions.
The energy-dependent creation of miRNAs is required for human miRNAs to perform the role that link seed regions from nucleotide positions 2-8 to microRNA flanking sequences linked to the recognition of cognate mRNA targets. The Creation of energy, and energy-dependent recognition allows the formation of intermolecular RNA-RNA base pair interactions that link the creation of miRNAs from the Creation of RNA to the light-activated formation of rG4 structures that link bindings with mRNA targets to biophysically constrained viral latency and healthy longevity via naturally occurring RNA interference in all living genera.
Simply put, we are what we eat in the context of transgenerational epigenetic inheritance across kingdoms.
See: Feedback loops link odor and pheromone signaling with reproduction (2005) and Mediterranean diet tied to lower risk of rheumatoid arthritis 9/11/18
Who will be next to address this link from light-activated microRNA biogenesis in plants to biophysically constrained food energy-dependent pheromone-controlled viral latency in humans?
For comparison, who will claim that they have discovered: A cell fitness selection model for neuronal survival during development?
There is only one model:
See:
Nutrient-dependent/pheromone-controlled adaptive evolution: a model (2013)
and the update:
Nutrient-dependent Pheromone-Controlled Ecological Adaptations: From Angstroms to Ecosystems (2018)