Trans-kingdom RNA interactions (2)
(click to enlarge)
The roles of structural dynamics in the cellular functions of RNAs 6/10/19
We discuss the mechanisms of gene regulation by microRNAs, riboswitches, ribozymes, post-transcriptional RNA modifications and RNA-binding proteins, and how the cellular environment and processes such as liquid–liquid phase separation may affect RNA folding and activity. The emerging RNA-ensemble–function paradigm is changing our perspective and understanding of RNA regulation, from in vitro to in vivo and from descriptive to predictive.
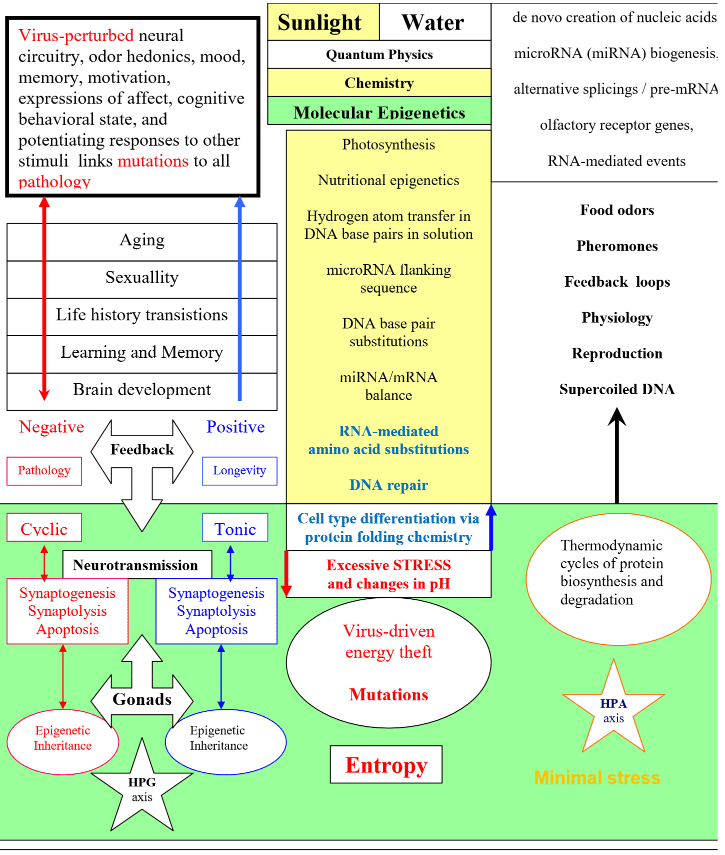
The creation of RNA is ATP-dependent. The Creation of sunlight and water has been predictably linked from Darwin’s “conditions of life” to all biodiversity on Earth via the physiology of reproduction. See: What is life when it is not protected from virus driven entropy (video)
See for comparison: Multigenerational epigenetic inheritance: One step forward, two generations back 8/27/19
Epigenetic patterns orchestrate gene expression programs that establish the phenotypic diversity of various cellular classes in the central nervous system, play a key role in experience-dependent gene regulation in the adult brain, and are commonly implicated in neurodevelopmental, psychiatric, and neurodegenerative disease states.
These authors claim that research that links trans-kingdom RNA interactions to multigenerational epigenetic inheritance in mammalian species is still in its infancy. See for comparison: From Fertilization to Adult Sexual Behavior (1996). In our Hormones and Behavior review we linked food energy-dependent alternative splicings of pre-mRNAs from the physiology of pheromone-controlled reproduction in yeasts to humans via the conserved molecular mechanisms of microRNA-mediated cell type differentiation. The term pre-mRNAs was changed to microRNAs, which helps to link our claims to everything known about cell type differentiation in all living genera to protection from the virus-driven degradation of messenger RNA that links mutations to all diseases.
See for comparison: Genomic Enhancers in Brain Health and Disease 1/14/19
Enhancers are non-coding DNA elements that function in cis to regulate transcription from nearby genes. Through direct interactions with gene promoters, enhancers give rise to spatially and temporally precise gene expression profiles in distinct cell or tissue types.
Pseudoscientists use terms like enhancers and promoters because they failed to link the energy-dependent creation of microRNAs from the light-activated assembly of the microRNA-RNA-peptide nanocomplex to biophysically constrained viral latency and the genesis of all biodiversity via the physiology of reproduction in species from microbes to humans.
In Genomic Enhancers in Brain Health and Disease 1/14/19 Energy-dependent gene regulation becomes enhancer-driven gene regulation.
See:

Regulation of gene expression patterns by genomic enhancers. Illustration of enhancer-promoter chromosomal looping (mediated by CTCF and cohesin) that allows distal enhancer elements to physically interact with and activate gene promoters. These interactions increase binding of transcription factors, chromatin modifiers, and the Mediator complex at gene promoters to recruit RNA polymerase II (RNAPII). Enhancers are characterized by elevated DNA sequence conservation, open chromatin, transcription factor binding motifs, characteristic histone modifications, DNA hypomethylation, and bidirectional transcription to generate enhancer RNAs (eRNAs).
The light-activated assembly of the microRNA-RNA-peptide nanocomplex becomes the mediator complex, which automagically links bidirectional transcription to the generation of enhancer RNAs (eRNAs).
In the world of biologically uninformed science idiots who fail to link the Creation of anti-entropic virucidal light and the Creation of water to the hydrophobicity of supercoiled DNA, pseudoscientists need only start with the generation of enhancer RNAs, to avoid the complexity of the ATP-dependent creation of RNA, which has linked the physiology of reproduction to the biogenesis of all diversity on Earth since 1964.
See: Dependence of RNA synthesis in isolated thymus nuclei on glycolysis, oxidative carbohydrate catabolism and a type of “oxidative phosphorylation”
The synthesis of RNA in isolated thymus nuclei is ATP dependent.
See also: UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond 1/17/19
Nonsense-mediated mRNA decay (NMD), which is arguably the best-characterized translation-dependent regulatory pathway in mammals, selectively degrades mRNAs as a means of post-transcriptional gene control. Control can be for the purpose of ensuring the quality of gene expression. Alternatively, control can facilitate the adaptation of cells to changes in their environment. Key to NMD, no matter what its purpose, is the ATP-dependent RNA helicase upstream frameshift 1 (UPF1), without which NMD fails to occur. However, UPF1 does much more than regulate NMD. As examples, UPF1 is engaged in functionally diverse mRNA decay pathways mediated by a variety of RNA-binding proteins that include staufen, stem-loop-binding protein, glucocorticoid receptor, and regnase 1. Moreover, UPF1 promotes tudor-staphylococcal/micrococcal-like nuclease-mediated microRNA decay. In this review, we first focus on how the NMD machinery recognizes an NMD target and triggers mRNA degradation. Next, we compare and contrast the mechanisms by which UPF1 functions in the decay of other mRNAs and also in microRNA decay. UPF1, as a protein polymath, engenders cells with the ability to shape their transcriptome in response to diverse biological and physiological needs.
Simply put, energy-dependent changes in the microRNA/messenger RNA balance must be linked to all biophysically constrained biodiversity via the physiology of reproduction, which biophysically constrains food energy-dependent pheromone-controlled trans-kingdom RNA interactions. For comparison, claims about enhancer-driven multigenerational epigenetic inheritance are ridiculous.
Want more on the same topic?
Swipe/Drag Left and Right To Browse Related Posts:
microRNA-mediated biodiversity (10)
3 MIN READ
0
RNAi 2002 to AI 2021 (1)
3 MIN READ
0
MicroRNA-mediated population control (10)
2 MIN READ
0
MicroRNA-mediated population control (6)
3 MIN READ
0
Quantum Darwinism (2)
3 MIN READ
0
Pheromones protect us from viruses (6)
5 MIN READ
0
God’s protection from SARS COV-2 and other viruses (1)
2 MIN READ
0
pH-dependent healthy longevity (2)
2 MIN READ
0
Transposons: crossroads, crosstalk and gene regulation
7 MIN READ
0
The tipping point (revisited): 97K (2)
3 MIN READ
0
Bruce McEwen’s legacy: sympatric speciation (4)
3 MIN READ
0
The tipping point (revisited): 96K (5)
4 MIN READ
0
The Darwin Code: Resurrected after 20 years (3)
2 MIN READ
0
microRNA-mediated prevention of diseases (1)
2 MIN READ
0
The tipping point (revisited): 95K (1)
< 1 MIN READ
0
MicroRNA-mediated, since 1964 (2)
3 MIN READ
0
Chemogenetic kinetics (3): Psychopathology
3 MIN READ
0
The tipping point (revisited): 91K (1)
3 MIN READ
0
Trans-kingdom RNA interactions (1)
2 MIN READ
0
Lewis Thomas (revisited): 90K (5)
4 MIN READ
0
Sunlight, hydrophobicity and biodiversity (2)
< 1 MIN READ
0
Relatable not debatable (2)
2 MIN READ
0
The tipping point (revisited): 87K (2)
3 MIN READ
0
The tipping point (revisited): 85,000 publications (4)
3 MIN READ
0
Kohl and Francoeur at 25 (3)
3 MIN READ
0
Biophysical constraints on RNA synthesis (3)
4 MIN READ
0
Darwin Day 2019 (3) #darwinday2019
3 MIN READ
0
Light-activated continuous environmental tracking (2)
3 MIN READ
0
The tipping point (revisited): 82,000 publications (1)
3 MIN READ
0
Light-activated morphogenesis and behavior (2)
4 MIN READ
0
From microRNA.pro to quantumsouls.pro (1)
4 MIN READ
0
Bottom-up biology (revisited)
3 MIN READ
0
Electrons build fractal shapes and prevent lung cancer
2 MIN READ
0
Evolved cancer prevention? (1)
2 MIN READ
0
The microRNA-mediated brain-microbiome axis
4 MIN READ
0
The concept of a species (3)
4 MIN READ
0
MicroRNA-mediated alternative splicing (revisited)
4 MIN READ
0
The eternal significance of microRNAs and the Vietnam Memorial (1)
3 MIN READ
0
Epigenetic inheritance of spatiotemporal regulation (3)
5 MIN READ
0
Epigenetic inheritance of spatiotemporal regulation (2)
5 MIN READ
0
Epigenetic inheritance of spatiotemporal regulation
4 MIN READ
0
A single base change refutes theistic evolution (2)
2 MIN READ
0
Ecological adaptations vs the randomness of evolution (3)
4 MIN READ
0
EDAR V370A and sympatric speciation
2 MIN READ
0
Virus-driven cystinosis
2 MIN READ
0
Environmental selection is natural selection (5)
5 MIN READ
0
Environmental selection is natural selection (4)
2 MIN READ
0
Environmental selection is natural selection (2)
2 MIN READ
0
MicroRNA-mediated denuclearization
2 MIN READ
0
Abiogenesis vs microRNA biogenesis
4 MIN READ
0
Let there be anti-entropic virucidal light
3 MIN READ
0
Part 3: Light-controlled cell biology (revisited)
3 MIN READ
0
Light-controlled cell biology (revisited)
6 MIN READ
0
Polymaths and paradigm shifts: From Asimov to Bear (5)
5 MIN READ
0
How to create biologically uninformed theorists
7 MIN READ
0
From photons to the proton motive force (2)
3 MIN READ
0
Ecological adaptation: a new definition of heredity (4)
4 MIN READ
0
Anti-entropic sunlight: Schrödinger’s Creationist Secret? (5)
2 MIN READ
0
Light-activated carbon fixation did not evolve (4)
7 MIN READ
0
The tipping point (revisited): 70,000 publications
4 MIN READ
0
George Church refutes theistic evolution (4)
4 MIN READ
0
Conceptual critique: Innateness vs the death gene (2)
5 MIN READ
0
Conceptual critique: Innateness vs the death gene (1)
5 MIN READ
0
Subatomic: From thermophiles to humans (3)
3 MIN READ
0
A reversible TCA cycle in a thermophile (3)
4 MIN READ
0
A mental health problem at the highest level (3)
7 MIN READ
0
Anti-entropic sunlight: Schrödinger’s Creationist Secret? (3)
6 MIN READ
1
The MicroRNAome Strikes Back: A Sokalian hoax (10)
3 MIN READ
0
Diet-driven RNA interference and mental health (2)
3 MIN READ
0
The MicroRNAome Strikes Back: A Sokalian hoax (4)
5 MIN READ
0
The MicroRNAome Strikes Back: A Sokalian hoax (3)
4 MIN READ
0
The MicroRNAome Strikes Back: A Sokalian hoax (1)
4 MIN READ
0
The tipping point (revisted): 69,000 publications
4 MIN READ
0
A mental health problem at the highest level (2)
3 MIN READ
0
Diet-driven RNA interference and cancer prevention (3)
6 MIN READ
0
Diet-driven RNA interference and cancer prevention
5 MIN READ
0
Interethnic genetic similarities and differences (2)
4 MIN READ
0
Quantized energy-dependent viral trophism (3)
3 MIN READ
0
The “walking fish” walks straight from quantum physics to quantum souls
6 MIN READ
0
From base editing to RNA editing (8)
4 MIN READ
0
Darwinian adaptation: a clear evolutionary trail?
4 MIN READ
0
Agilent technology and energy-dependent autophagy
8 MIN READ
0
Elsevier fails to support the concept of autophagy
4 MIN READ
0
From Autophagy.pro (transitions)
4 MIN READ
0
Nature vs Science and Autophagy.pro
7 MIN READ
0
The overwhelming ignorance of sex researchers
5 MIN READ
0
Who created your virus-driven death gene? (1)
5 MIN READ
0
Pheromones biophysically constrain RNA-mediated biodiversity (1)
5 MIN READ
0
Pheromones biophysically constrain base editing and RNA editing
3 MIN READ
0
From base editing to RNA editing (2)
7 MIN READ
0
Cryo-EM: More than a suggestion
< 1 MIN READ
0
Losers: rediscovering an epigenetic key
2 MIN READ
0
Virulence: Out of stock
2 MIN READ
0
Energy-dependent structure and function: Until death (5)
4 MIN READ
0
Energy-dependent structure and function: Until death (3)
3 MIN READ
0
Energy-dependent structure and function: Until death (2)
5 MIN READ
0
Energy-dependent structure and function: Until death (1)
4 MIN READ
0
Predicting who wins the 2017 Nobel Prizes (7)
4 MIN READ
0
Until death: Virus-driven failure of multisensory integration (1)
4 MIN READ
0
Predicting who wins the 2017 Nobel Prizes (6)
5 MIN READ
0
Predicting who wins the 2017 Nobel Prizes
4 MIN READ
0
Caught in an epigenetic trap: flag waving consequences
< 1 MIN READ
0
Life and death via chemical effects on photosynthesis
8 MIN READ
0
Inventing “Transcriptome Trajectory Turning Points”
2 MIN READ
0
Sexual communication signals: New Insights!
10 MIN READ
0
Quantum physics, Patrick Bateson, and neo-Darwinian nonsense
3 MIN READ
0
Neurosteroids revisited two decades later
3 MIN READ
0
Host-derived creation of all pathology (2 of 2)
9 MIN READ
0
God vs host-derived creation of virus-driven pathology (2)
4 MIN READ
0
God vs host-derived creation of virus-driven pathology
6 MIN READ
0
Pseudoscientists hate what science explains! (3)
4 MIN READ
0
Your indifference is killing you and others
5 MIN READ
0
Robert Sapolsky’s legacy of atheistic pseudoscientific nonsense
4 MIN READ
0
Energy-dependent physical and biophysical constraints (8)
5 MIN READ
0
Energy-dependent physical and biophysical constraints (7)
2 MIN READ
0
Energy-dependent physical and biophysical constraints (5)
7 MIN READ
0
Energy-dependent physical and biophysical constraints (4)
2 MIN READ
0
Energy-dependent physical and biophysical constraints (2)
4 MIN READ
0
Richard P. Feynman refuted theistic evolution
5 MIN READ
0
God’s shrinking role in salvation (2)
5 MIN READ
0
Food energy-dependent cell type differentiation (2)
10 MIN READ
0
Hard Science vs the neo-Darwinian “magic box”
9 MIN READ
0
Food energy-dependent epigenetic adaptation
11 MIN READ
0
Energy-dependent epigenetic translation to mRNA stability (6)
8 MIN READ
0
Energy-dependent epigenetic translation to mRNA stability (5)
4 MIN READ
0
Nothing evolved into all biodiversity on Earth
4 MIN READ
0
Energy as information and constrained endogenous RNA interference (4)
7 MIN READ
0
The death of human ethology via ecology
7 MIN READ
0
Dispensing with all pseudoscientific nonsense about evolution (2)
< 1 MIN READ
0
Dispensing with March for Science Dispatches (2)
2 MIN READ
0
Virus-driven downsizing of the human brain (4)
2 MIN READ
0
March for science vs microRNAs
3 MIN READ
0
Open Science: Closed to facts about microRNAs
6 MIN READ
0
Viruses in pathogenic variants disrupt alternative splicings
2 MIN READ
0
Epigenetically-effected nucleosome repositioning sheds Dobzhansky’s light on evolution
3 MIN READ
0
Synthetic RNA-based switches
2 MIN READ
0
Sunlight bursts the social bubble of physics and math
3 MIN READ
0
Functionally interdependent editing and methylation
4 MIN READ
0
Light-activated feedback loops vs self-organization of ecosystems
7 MIN READ
0
Harvard researchers support young earth creationism
< 1 MIN READ
0
Cytosis: A Cell Biology Board Game
< 1 MIN READ
0
Life in your UV light-constrained galaxy
5 MIN READ
0
Food for thought: Penrose v Ellis (1)
5 MIN READ
0
Proof: Sunlight is energy as information
< 1 MIN READ
0
Energy-dependent pheromone-controlled entropy (2)
6 MIN READ
0
The essence of precision medicine: drug targets or healthy longevity?
4 MIN READ
0
RNA-mediated adult learning, memory, and neurogenesis
3 MIN READ
0
Theistic evolutionists fight back and lose (2)
4 MIN READ
0
Young earth creationists refute theistic evolution
3 MIN READ
0
Charles Darwin refutes theistic evolution
2 MIN READ
0
Emily Witkin refutes theistic evolution
2 MIN READ
0
Serious scientists refute theistic evolution again
3 MIN READ
0
Pseudoscientists fail to refute theistic evolution
4 MIN READ
0
May the anti-entropic force of sunlight be with you
4 MIN READ
0
Energy is information. Objections over ruled. You’re fired!
7 MIN READ
0
Happy Darwin Day (2017)
4 MIN READ
0
George Church refutes theistic evolution (3)
5 MIN READ
0
Fighting for virus-driven pathology
5 MIN READ
0
ATP-dependent creation of RNA
2 MIN READ
0
Dobzhansky 1973 and Precision Medicine (2)
5 MIN READ
0
Science journalists or paid propagandists? (4)
6 MIN READ
0
Theorists sell hidden energy
3 MIN READ
0
Energy-dependent chirality
6 MIN READ
0
Energy-dependent alternative splicings 1996 – 2016 (2)
5 MIN READ
0
Vietnam Veterans and others with glioblastoma
3 MIN READ
0
Energy-dependent sensory maps (1996-2016)
5 MIN READ
0
Energy-dependent oscillating gene networks organize life
4 MIN READ
0
2016 obfuscated facts about energy as information
5 MIN READ
0
More refutations of neo-Darwinian nonsense
7 MIN READ
0
Re-inventing mutation-driven evolution
3 MIN READ
0
Sudden death indel polymorphism
4 MIN READ
0
Anti-entropic virucidal energy as information
10 MIN READ
0
Tasting light links energy from creation to adaptation
9 MIN READ
0
Politicized science: The demise of RNA-mediated.com?
3 MIN READ
0
Combating evolution: Battlefield medicine vs politicized science
6 MIN READ
0
Metabolic Phenotyping Research
2 MIN READ
0
Energy-dependent coulombic, autophagic, polycombic healthy longevity
4 MIN READ
0
Energy-dependent purifying selection / autophagy (2)
2 MIN READ
0
Base pairs, amino acids and phenotypes
10 MIN READ
0
Building delicious base pairs in class. It’s elementary Watson and Crick
3 MIN READ
0
Building delicious base pairs in class. It's elementary Watson and Crick
3 MIN READ
0
Virus-driven mutation or amino acid substitution
5 MIN READ
0
Epigenetically effected energy-dependent fluorescence
5 MIN READ
0
Epigenetics and autophagy vs mutations and evolution (7)
3 MIN READ
0
Top-down adaptation vs bottom-up evolution
6 MIN READ
0
Light ‘drives’ adaptation; nothing ‘drives’ evolution (3)
< 1 MIN READ
0
Polycombic ecological adaptation as a science, not a theory (2)
12 MIN READ
0
Polycombic ecological adaptation as a science, not a theory
3 MIN READ
0
Attacking Young Earth Creationists
5 MIN READ
0
80 years of causal analysis (1936 – 2016)
11 MIN READ
0
Carl Woese was wrong
2 MIN READ
0
The natural success of RNAi and failed treatment
7 MIN READ
0
Did evolution autophosphorylate your kinases? (2)
5 MIN READ
1
Nutrient-dependent autophagy
3 MIN READ
0
Life is energy-dependent task management
3 MIN READ
0
Hypothesis free pseudoscience vs facts (5)
6 MIN READ
0
Hypothesis free pseudoscience vs facts (4)
5 MIN READ
0
The last RNA-mediated theory killer
< 1 MIN READ
0
Chromatin: The structure of DNA (3)
4 MIN READ
0
Chromatin: The structure of DNA
6 MIN READ
0
DNA repair via junk DNA (2)
5 MIN READ
0
DNA repair via junk DNA (1)
5 MIN READ
0
Linking RNA structure to function
3 MIN READ
0
Increased soil pH and nutrient availability
4 MIN READ
0
Lawless evolution: No physics
4 MIN READ
0
Anthetical conclusions (2)
5 MIN READ
0
RNAi: From magic bullet to billion dollar baby
4 MIN READ
0
The end of neo-Darwinism
5 MIN READ
0
Biophotonics, glycobiology, quantized biodiversity
4 MIN READ
0
The Origin of Information (2)
6 MIN READ
0
Did “Nature” kill Steve Jobs? (3)
10 MIN READ
0
Funding the Human Genome Project-Write
2 MIN READ
0
Virus-driven downsizing of the human brain
2 MIN READ
0
MicroRNAs and sexual orientation
8 MIN READ
0
Major transition ends use of silly theories
5 MIN READ
0
Masters of deception about nature (2)
7 MIN READ
0
Energy-dependent RNA methylation (2)
5 MIN READ
0
War Games: False Flag Terrorism
5 MIN READ
0
Countdown to Genetics and Genomics
< 1 MIN READ
0
Wasted Templeton Funding (4)
3 MIN READ
0
RNA splicing, genetic variation, and disease
2 MIN READ
0
Creating and/or programming the immune system
< 1 MIN READ
0
Energy dependent RNA-mediated immunity (3)
8 MIN READ
0
Effects on invertebrate GnRH and affects on primate behavior
6 MIN READ
0
Creating gravity, nucleic acids, receptors, and supercoiled DNA (2)
9 MIN READ
0
Ricki Lewis’ Time Machine
5 MIN READ
0
Hydrogen-atom transfer in DNA base pairs (5)
10 MIN READ
0
The radical pair mechanism of ecological speciation
3 MIN READ
0
Ecological genomics: teleophobes respond (too late)
10 MIN READ
0
Genes, orchid odors, and pheromones from blonds
5 MIN READ
0
RNA methylation, RNA-directed DNA methylation, learning and memory
3 MIN READ
0
Finding peace and π in the light of H bond energy
5 MIN READ
0
Epigenetic (re)programming of behavior (2)
6 MIN READ
0
Life history transitions and RNA-mediated survial
2 MIN READ
0
Does metabolism link beneficial mutations to cancer?
10 MIN READ
0
A two-faced protein enables RNA-mediated DNA repair (2)
4 MIN READ
0
Teaching the biologically uninformed
3 MIN READ
0
Stress-perturbed mitochondrial dysfunction (2)
5 MIN READ
0
Too complex for the Complex Biological Systems Alliance
4 MIN READ
0
Conserved molecular mechanisms
< 1 MIN READ
0
Models are not theories (2)
2 MIN READ
0
Mechanisms of stress: from genes to cancer
9 MIN READ
0
The stability of organized genomes
5 MIN READ
0
MicroRNA controlled growth and brain development
7 MIN READ
0
MicroRNA – controlled ecological adaptations
5 MIN READ
0
MicroRNAs and the exposome (2)
< 1 MIN READ
0
DNA Methylation and organized genomes
7 MIN READ
0
Amino acid-dependent cell type differentiation
8 MIN READ
0
Origin of life and cancer (1,2,3)
2 MIN READ
0
The miRNA/mRNA balance: a suboptimal strategy?
2 MIN READ
0
Too many targets for theories
3 MIN READ
0
Anti-entropic solar energy
5 MIN READ
0
Mimicking claims and ignoring facts
4 MIN READ
0
Rejecting what is known about viral microRNAs and nutrient-dependent microRNAs
2 MIN READ
0
Communication, not mutations
2 MIN READ
0
Biologists puzzled by evolved RNAs and decaying DNA
6 MIN READ
0
Complex behaviors of cell types in cancer
6 MIN READ
0
RNA-mediated cell type differentiation and behavior
3 MIN READ
0
Memory of repression and memory of behavior
2 MIN READ
0
Pattern recognition and conserved receptors (TAARs)
5 MIN READ
0
Ecologically linked adapted ants and brains
2 MIN READ
0



